Recently, new advanced high-throughput sequencing technology as a novel tool has opened the way to study of genomic variants and functional information stored within farm animals.
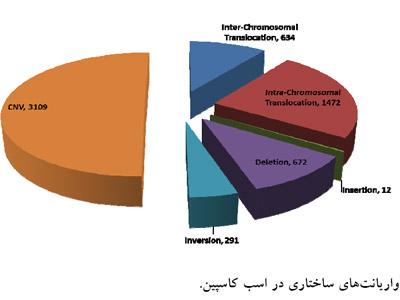
The Caspian horse is one of the valuable horses ever exist in the world. Hence, propose of this study was to investigate genetic variants of single nucleotide polymorphisms, insertion and deletions and copy number variations within the genome of Caspian horse and their involved biological pathways. Using high-throughput sequencing technology, we generated 108 Gb (Average depth of 45.8) of DNA sequence from three Caspian horse mares resulting in an average of 14.41X coverage and 76.4% covered with reference genome. Using a stringent filtering method, we identified 1666717 single nucleotide polymorphisms, 358020 insertion and deletions, and 3109 copy number variations. Functional clustering analysis of genic variants revealed that most of the genetic variants in the Caspian horse’s genome were enriched in nervous system, GTP-related signal transduction, cellular morphogenesis, cytoskeleton organization, vascular development and cellular movement. Moreover, we have detected structural variations as like as inversion, intra- and inter-chromosomal translocations, large insertion and deletions which could be useful for marker based population genetic investigation.
Keywords, High-throughput sequencing, Caspian horse, Genic variants, Biological pathways.
Arefnezhad B.1, Kohram H.1, Moradi Shahre-Babak M.1, Shakeri M.1, Dong Y.5, Zhang X.5, Wang W.5, Hoseini Salekdeh Gh.*4
1Department of Animal Science, University of Tehran, Karaj, Iran.
2 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China.
3Agricultural Biotechnology Research Institute of Iran (ABRII), Karaj, Iran.
*Corresponding Author: Kohram H, Hoseini Salekdeh Gh. Tel: 02632248082 Email: hamid.kohram@yahoo.com, hsalekdeh@yahoo.com
references:
Abyzov A, Urban AE, Snyder M, Gerstein M (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Research 21: 974-984.
Andrews S (2012). FASTQC. A quality control tool for high throughput sequence data. URL http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP (2009). BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods 6: 677-681.
Cingolani P, Platts A, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80-92.
Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA (2008). Identification of genetic variants using bar-coded multiplexed sequencing. Nature Methods 5: 887-893.
Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T (2008). Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Research 36: e122-e122.
Dennis Jr G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biology 4: P3.
Doan R, Cohen ND, Sawyer J, Ghaffari N, Johnson CD, Dindot SV (2012). Whole-Genome sequencing and genetic variant analysis of a quarter Horse mare. BMC Genomics 13: 78.
Firouz L (1969). Conservation of a domestic breed. Biological Conservation 2: 53-54.
Firouz L (1971). Osteological and historical implication of the Caspian pony to early domestication in Iran. Proc 3rd Int Congr Agricultural Museum, Budapest: 1-5.
Firouz L (1972). The Caspian miniature horse of Iran. Field Research Projects, Florida, USA,
Garrison E, Marth G (2012). Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:12073907.
Hatami-Monazah H, Pandit RV (1979). A cytogenetic study of the Caspian pony. Journal of Reproduction and Fertility 57: 331-333.
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T (2009). High-throughput genotyping by whole-genome resequencing. Genome Research 19: 1068-1076.
Langmead B (2002). Aligning Short Sequencing Reads with Bowtie. Current Protocols in Bioinformatics: John Wiley & Sons, Inc.
Lindgreen S (2012). AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Research Notes 5: 337.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20: 1297-1303.
Orlando L, Ginolhac Al, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I (2013). Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499: 74-81
Shahsavarani H, Rahimi-Mianji G (2012). Analysis of genetic diversity and estimation of inbreeding coefficient within Caspian horse population using microsatellite markers. African Journal of Biotechnology 9: 293-299
Shao H, Bellos E, Yin H, Liu X, Zou J, Li Y, Wang J, Coin LJM (2012). A population model for genotyping indels from next-generation sequence data. Nucleic acids research 41: e46-e46.
Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blöcker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MCT, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guérin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Røed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvänen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Broad Institute Genome Sequencing P, Broad Institute Whole Genome Assembly T, Lander ES, Lindblad-Toh K (2009). Genome Sequence, Comparative Analysis, and Population Genetics of the Domestic Horse. Science 326: 865-867.
Wang K, Li M, Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research 38: e164-e164.
Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ (1998). Direct allelic variation scanning of the yeast genome. Science 281: 1194-1197.