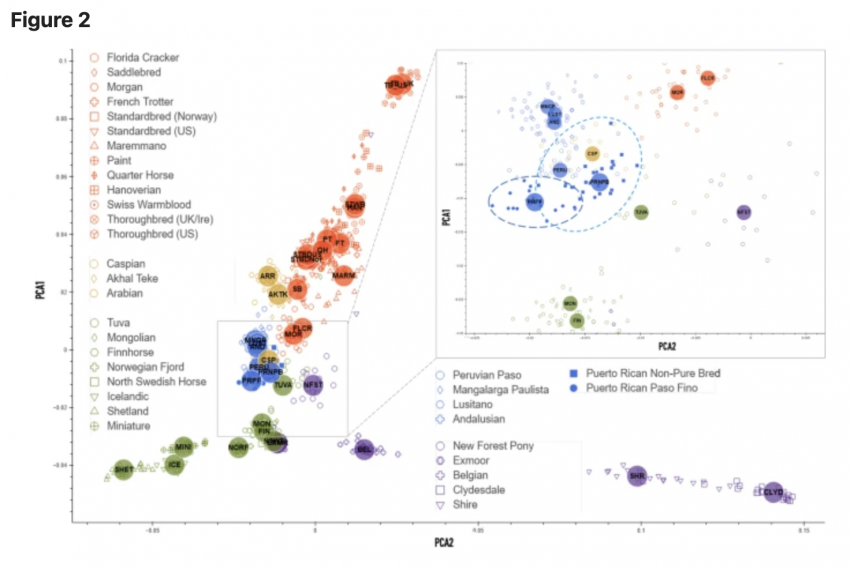
In particular, PRNPB horses share genetic diversity components with New Forest Pony, Tuva, Mongolian and Caspian horses and Florida Cracker.
Scientifc Reports | (2022) 12:515 | https://doi.org/10.1038/s41598-021-04537-5
Walter W. Wolfsberger1,2,3,7, Nikole M.Ayala2,7, Stephanie O. Castro‑Marquez1,2, Valerie M. Irizarry‑Negron2 , Antoliy Potapchuk2 , Khrystyna Shchubelka1,2,3, Ludvig Potish4 , Audrey J. Majeske1,2, Luis Figueroa Oliver2 , Alondra Diaz Lameiro2 , Juan Carlos Martínez‑Cruzado2 , Gabriella Lindgren5,6 & Taras K. Oleksyk1,2,3*
Abstract
Since the first Spanish settlers brought horses to America centuries ago, several local varieties and breeds have been established in the New World. These were generally a consequence of the admixture of the different breeds arriving from Europe. In some instances, local horses have been selectively bred for specific traits, such as appearance, endurance, strength, and gait. We looked at the genetics of two breeds, the Puerto Rican Non-Purebred (PRNPB) (also known as the “Criollo”) horses and the Puerto Rican Paso Fino (PRPF), from the Caribbean Island of Puerto Rico. While it is reasonable to assume that there was a historic connection between the two, the genetic link between them has never been established. In our study, we started by looking at the genetic ancestry and diversity of current Puerto Rican horse populations using a 668 bp fragment of the mitochondrial DNA D-loop (HVR1) in 200 horses from 27 locations on the island. We then genotyped all 200 horses in our sample for the “gait-keeper” DMRT3 mutant allele previously associated with the paso gait especially cherished in this island breed. We also genotyped a subset of 24 samples with the Illumina Neogen Equine Community genome-wide array (65,000 SNPs). This data was further combined with the publicly available PRPF genomes from other studies. Our analysis show an undeniable genetic connection between the two varieties in Puerto Rico, consistent with the hypothesis that PRNPB horses represent the descendants of the original genetic pool, a mix of horses imported from the Iberian Peninsula and elsewhere in Europe. Some of the original founders of PRNRB population must have carried the “gait-keeper” DMRT3 allele upon arrival to the island. From this admixture, the desired traits were selected by the local people over the span of centuries. We propose that the frequency of the mutant “gait-keeper” allele originally increased in the local horses due to the selection for the smooth ride and other characters, long before the PRPF breed was established. To support this hypothesis, we demonstrate that PRNPB horses, and not the purebred PRPF, carry a signature of selection in the genomic region containing the DMRT3 locus to this day. The lack of the detectable signature of selection associated with the DMRT3 in the PRPF would be expected if this native breed was originally derived from the genetic pool of PRNPB horses established earlier and most of the founders already had the mutant allele. Consequently, selection specific to PRPF later focused on allels in other genes (including CHRM5, CYP2E1, MYH7, SRSF1, PAM, PRN and others) that have not been previously associated with the prized paso gait phenotype in Puerto Rico or anywhere else.
Genome diversity, structure and admixture
The genome-wide diversity of the two horse breeds was evaluated with the Illumina Neogen Equine Community Array containing 65,157 markers12. Then, using an overlap between genome-wide genotyping arrays, we increased our sample set of PRPFs by adding publicly available data from a broader study based on similar technology13. The two largest components (PC1 and PC2) from the principal components analysis (PCA) were then matched to all horse samples across of the studies to separate the Puerto Rican horses from other American breeds. The resulting distribution shows genetic differences between Puerto Rican horses and all other breeds (Fig. 2), which can be clearly distinguished: the two breeds (PRPF and PRNPB) form a distinct spread along a “vector” (shown by the oval in the magnified insert in Fig. 2), with PRNPB occupying the proximal region and PRPF at the distal region of the spread. The three Iberian breeds that occupy the same branch on the phylogenetic tree (Lusitano, Andalusian and Mangalanga Paulista, group “C”, Fig. S1) are clustered nearby, as expected but, horses from other branches seem to cluster even closer, specifically, the Caspian and Tuvan horses. While some of the diversity in the nonpurebred NPB Puerto Rican Criollo horses is also shared with the Peruvian Paso, the PRPF horse genotypes form a clearly distinct cluster, distinctive from all other breeds (Fig. 2).
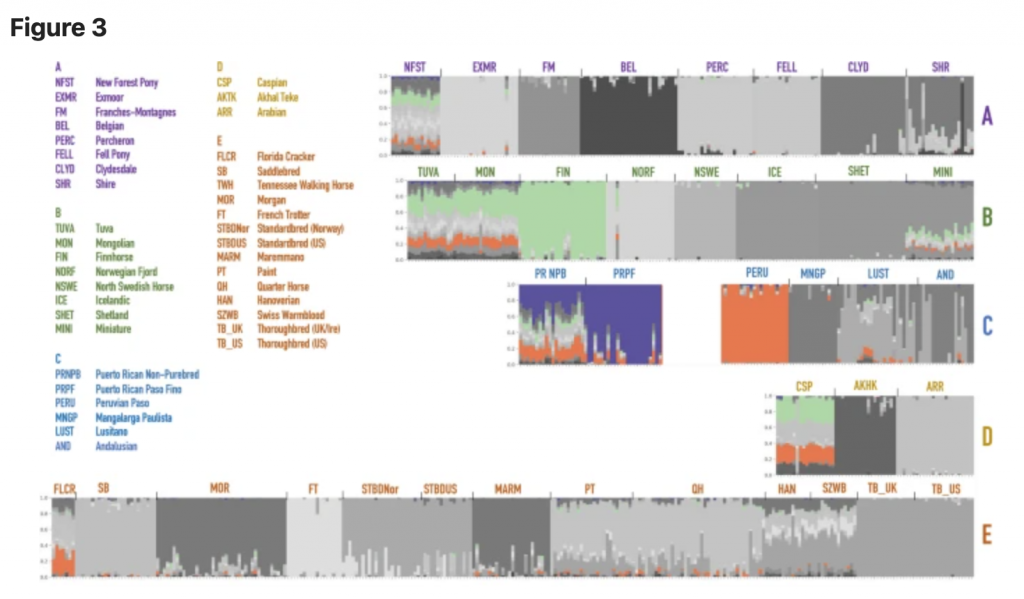
The admixture analysis performed on the merged datasets identified genetic components that may be shared between the two Puerto Rican horses and other breeds. While the original visualization of all horse breeds produceed a hard to read cluttered plot, we highlighted segments14 that were shared between different breeds (Fig. 3). For convenience, the component colors were preserved from the structure analysis in earlier studies13, and the new plot was separated into five larger groups based on the neighbor-joining tree calculated from SNP frequencies in 38 horse populations in Petersen et al.13 (Fig. S1). According to the structure and admixture analysis, Puerto Rican horses share population structure components with a number of horses worldwide (Fig. 3). In particular, PRNPB horses share genetic diversity components with New Forest Pony, Tuva, Mongolian and Caspian horses and Florida Cracker. All of these horses show multiple genetic components (shown by the presence of different colors in the plot), which indicate diversity within the breeds. On the other hand, the PRPF genome is largely dominated by a single (purple) component that seems to be almost entirely unique to this breed. However, traces of this component can also be seen in New Forest Pony, Tuva, and Mongolian horses (Fig. 3). Interestingly, whie the Peruvian Paso also shares a genetic component with PRNPB, it seems to be dominated by a different (orange), more common component than that which is prevalent in the PRPF.
Discussion
Thanks to the analysis of the genome-wide array data, we can see that Puerto Rican horses share genome variation components with a number of horses worldwide (Fig. 3). In particular, the PRNPB horses appear to have genomic fragments in common with the Northern European and Asian horse breeds (Fig. 3, top row). Specifically, they share the “light green” and the “orange” components with the Finnhorses, Mongolian and Tuvan breeds. This appears to be the same component present in the Iberian (Lusitano), Middle Eastern (Caspian horse), or US derivatives from the Spanish stock brought to Florida in the 1500s (Florida Cracker). The “orange” component present on the island, also completely dominates the Peruvian Paso, the breed that is most closely related to the PRNPB horses outside of Puerto Rico. Both Puerto Rican breeds display a common “purple” component that seems to be unique to the local island horses and cannot be found in any of the surveyed horse breeds at the time (Fig. 3). This component represents a larger part of genetic variation in the PRNPB horse (which also has green and orange components shared with other breeds) but completely dominates the PRPF genomes. The most likely explanation of this observation is that the PRNPB horse has a unique mixture that incorporates variation from a diverse set of lines brought on the ships to the island, and the PRPF has been selected for this particular set of variation from the admixed pool. If the latter statement is true, PRPF should have less genetic diversity than PRNPB.